
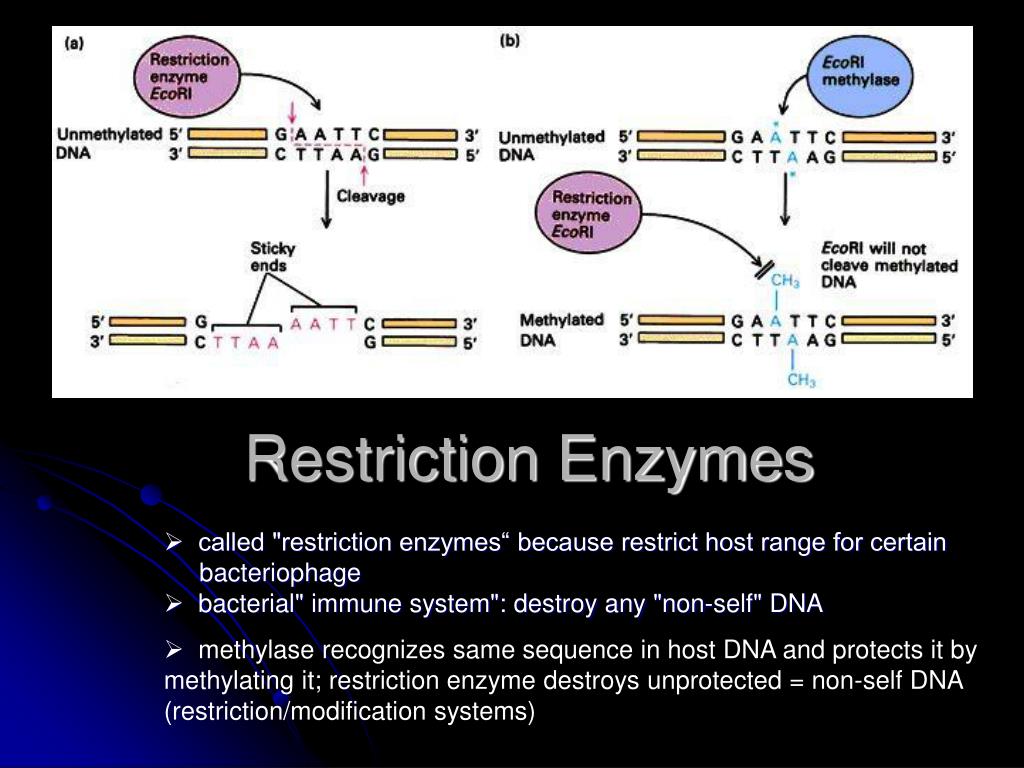
To be able to sequence DNA, it is first necessary to cut it into smaller fragments. The recognition sites of restriction enzymes are often palindromic, meaning that the DNA sequence reads the same in both forward and reverse directions. the recognition site (also known as a restriction site). REs occur naturally in bacteria, where they specifically recognize short stretches of nucleotides in DNA and catalyze double-strand breaks at or near. Because they cut within the molecule, they are often called restriction endonucleases. The discovery of restriction enzymes, or restriction endonucleases (REs), was pivotal to the development of molecular cloning. A few possible pathways are discussed for MspI to cut both strands of DNA, either as a monomer or dimer.ĭepartment of Physiology and Biophysics, Boston University School of Medicine, 715 Albany Street, MA 02118, USA. Restriction enzymes are DNA-cutting enzymes found in bacteria (and harvested from them for use). This MspI-DNA structure represents the first example of asymmetric recognition of a palindromic DNA sequence by two different structural motifs in one polypeptide. The recognition sequences of Type II restriction enzymes are usually inverted repeats, so that the enzyme cuts between the same bases on both strands. The enzyme makes specific contacts with all 4 base pairs in the recognition sequence, by six direct and five water-mediated hydrogen bonds and numerous van der Waal contacts. Unlike any other Type IIP enzyme reported to date, an MspI monomer and not a dimer binds to a palindromic DNA sequence. Here we report an asymmetric complex of the Type IIP restriction enzyme MspI in complex with its cognate recognition sequence. Due to the recognition and cleavage symmetry, Type IIP enzymes are usually found to act as homodimers in forming 2-fold symmetric enzyme-DNA complexes. Most well-known restriction endonucleases recognize palindromic DNA sequences and are classified as Type IIP. Diversity, Equity, Inclusion, and Access.The restriction enzyme sites in the right places relative to each other b) Why does lane 1 have 2 bands in it (3 points) Uncut plasmid will migrate in an agarose gel based on its shape as well as its size. Citation, Usage, Privacy Policies, Logo The following is part of the sequence of an 8 base pair, palindromic restriction enzyme recognition site.Biologically Interesting Molecule Reference Dictionary (BIRD).


 0 kommentar(er)
0 kommentar(er)
